I am changing to GFS 0.25 to feed my WRF models (about time). I decided to install WRF 4.0.2 as I knew prior versions to 3.9.0.1 had a problem giving Soil temperatures of 0 at some points. I compiled the new model using intel compilers (and intel compiled netcdf libraries).
I tested the new 4.0.2 model and everything went fine but when wrf.exe started, it instantaneously crashed giving a segmentation fault (and no extra info). I attach the last part of the rsl.error (the ones that had an error).
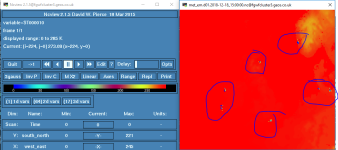
First thing I checked is that the met_em files had the right temperature, and bingo! the soil temperatures still had zero values on some points.
I have tried compiling and running WPS versions 3.9.1 and 3.9.0.1 of WRF to see if that solved the problem but it doesn't.
in case it may be of use I run WRF in a AWS cluster using a C4.x instance with 36 cores (that worked fine for the other version of WRF, 3.6.1).
METGRID.TBL.AWR and Vtable.GFS are the correct ones (the exact same file for the three versions, 4.02,3.9.1,3.9.0.1, the WPSv3.6.1 gave NUM_MET_SOIL_LEV =0 so I had to change)
-----------------------------------------------
rsl.error.00xx (only the tail part):
-----------------------------------------------
INITIALIZE THREE Noah LSM RELATED TABLES
mediation_integrate.G 1943 DATASET=HISTORY
mediation_integrate.G 1944 grid%id 1 grid%oid 1
d01 2018-12-12_00:00:00 Input data is acceptable to use:
d01 2018-12-12_00:00:00 Input data processed for aux input 10 for domain 1
d01 2018-12-12_00:00:00 Input data is acceptable to use:
Tile Strategy is not specified. Assuming 1D-Y
WRF TILE 1 IS 105 IE 156 JS 95 JE 140
WRF NUMBER OF TILES = 1
forrtl: severe (174): SIGSEGV, segmentation fault occurred
Image PC Routine Line Source
wrf.exe 000000000303DE25 Unknown Unknown Unknown
wrf.exe 000000000303BBE7 Unknown Unknown Unknown
wrf.exe 0000000002FDD184 Unknown Unknown Unknown
wrf.exe 0000000002FDCF96 Unknown Unknown Unknown
wrf.exe 0000000002F69026 Unknown Unknown Unknown
wrf.exe 0000000002F6FB00 Unknown Unknown Unknown
libpthread.so.0 00001476F82695E0 Unknown Unknown Unknown
wrf.exe 00000000028F7FA3 Unknown Unknown Unknown
wrf.exe 00000000028F1177 Unknown Unknown Unknown
wrf.exe 00000000023A7A3D Unknown Unknown Unknown
wrf.exe 0000000001BA992F Unknown Unknown Unknown
wrf.exe 0000000001319588 Unknown Unknown Unknown
wrf.exe 00000000011A2570 Unknown Unknown Unknown
wrf.exe 000000000050EB8F Unknown Unknown Unknown
wrf.exe 000000000040C391 Unknown Unknown Unknown
wrf.exe 000000000040C34F Unknown Unknown Unknown
wrf.exe 000000000040C2EE Unknown Unknown Unknown
libc.so.6 00001476F7BAD445 Unknown Unknown Unknown
wrf.exe 000000000040C1E9 Unknown Unknown Unknown
I tested the new 4.0.2 model and everything went fine but when wrf.exe started, it instantaneously crashed giving a segmentation fault (and no extra info). I attach the last part of the rsl.error (the ones that had an error).
First thing I checked is that the met_em files had the right temperature, and bingo! the soil temperatures still had zero values on some points.
I have tried compiling and running WPS versions 3.9.1 and 3.9.0.1 of WRF to see if that solved the problem but it doesn't.
in case it may be of use I run WRF in a AWS cluster using a C4.x instance with 36 cores (that worked fine for the other version of WRF, 3.6.1).
METGRID.TBL.AWR and Vtable.GFS are the correct ones (the exact same file for the three versions, 4.02,3.9.1,3.9.0.1, the WPSv3.6.1 gave NUM_MET_SOIL_LEV =0 so I had to change)
-----------------------------------------------
rsl.error.00xx (only the tail part):
-----------------------------------------------
INITIALIZE THREE Noah LSM RELATED TABLES
mediation_integrate.G 1943 DATASET=HISTORY
mediation_integrate.G 1944 grid%id 1 grid%oid 1
d01 2018-12-12_00:00:00 Input data is acceptable to use:
d01 2018-12-12_00:00:00 Input data processed for aux input 10 for domain 1
d01 2018-12-12_00:00:00 Input data is acceptable to use:
Tile Strategy is not specified. Assuming 1D-Y
WRF TILE 1 IS 105 IE 156 JS 95 JE 140
WRF NUMBER OF TILES = 1
forrtl: severe (174): SIGSEGV, segmentation fault occurred
Image PC Routine Line Source
wrf.exe 000000000303DE25 Unknown Unknown Unknown
wrf.exe 000000000303BBE7 Unknown Unknown Unknown
wrf.exe 0000000002FDD184 Unknown Unknown Unknown
wrf.exe 0000000002FDCF96 Unknown Unknown Unknown
wrf.exe 0000000002F69026 Unknown Unknown Unknown
wrf.exe 0000000002F6FB00 Unknown Unknown Unknown
libpthread.so.0 00001476F82695E0 Unknown Unknown Unknown
wrf.exe 00000000028F7FA3 Unknown Unknown Unknown
wrf.exe 00000000028F1177 Unknown Unknown Unknown
wrf.exe 00000000023A7A3D Unknown Unknown Unknown
wrf.exe 0000000001BA992F Unknown Unknown Unknown
wrf.exe 0000000001319588 Unknown Unknown Unknown
wrf.exe 00000000011A2570 Unknown Unknown Unknown
wrf.exe 000000000050EB8F Unknown Unknown Unknown
wrf.exe 000000000040C391 Unknown Unknown Unknown
wrf.exe 000000000040C34F Unknown Unknown Unknown
wrf.exe 000000000040C2EE Unknown Unknown Unknown
libc.so.6 00001476F7BAD445 Unknown Unknown Unknown
wrf.exe 000000000040C1E9 Unknown Unknown Unknown