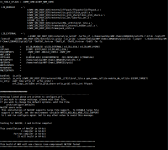
Hi, I am getting this error with WRF 4.2. There are similar threads in the forum but no exact solution.
WRF was installed successfully. I have tried the same steps in installation as with previous versions of WRF (4.1.3) and I never faced such an error.Also large file support was enabled and sufficient disk space is there.
Kindly help!
Also
When configuring WRF-4.1.3 , This build of WRF will use classic (non-compressed) NETCDF format is shown. 4.1.3 is working properly.
While configuring WRF-4.2, This build of WRF will use NETCDF4 with HDF5 compression is shown.
Both are configured with the same libraries of NETCDF4 and HDF5. I am confused why is the difference. Could it be a reason for the above error?
Also tried tried different HDF5 versions if it matters.
WRF was installed successfully. I have tried the same steps in installation as with previous versions of WRF (4.1.3) and I never faced such an error.Also large file support was enabled and sufficient disk space is there.
Kindly help!
Also
When configuring WRF-4.1.3 , This build of WRF will use classic (non-compressed) NETCDF format is shown. 4.1.3 is working properly.
While configuring WRF-4.2, This build of WRF will use NETCDF4 with HDF5 compression is shown.
Both are configured with the same libraries of NETCDF4 and HDF5. I am confused why is the difference. Could it be a reason for the above error?
Also tried tried different HDF5 versions if it matters.