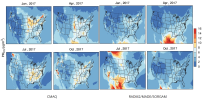
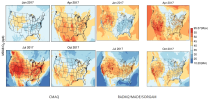
Hi all,
I did a hindcast on CONUS for PM2.5 and ozone with RADM2/MADE/SORGAM using WRF/Chem 3.9.1.1. I compared my simulation result with CMAQ and EPA observations. My ozone simulation looks reasonable, while PM2.5 is different with both CMAQ results and EPA observation of regional distribution and order of magnitude.
May I ask for help/comments on my process?
1, I used NEI 2017 and processed it with the EPA_ANTHRO_EMIS tool. Is there any modification needed in my "EPA_anthro_emis_2017NEI.inp"?
2, Did anyone get good PM2.5 CONUS simulation results with RADM2/MADE/SORGAM?
3, I appreciate any other suggestions.
Best,
Phillip
I did a hindcast on CONUS for PM2.5 and ozone with RADM2/MADE/SORGAM using WRF/Chem 3.9.1.1. I compared my simulation result with CMAQ and EPA observations. My ozone simulation looks reasonable, while PM2.5 is different with both CMAQ results and EPA observation of regional distribution and order of magnitude.
May I ask for help/comments on my process?
1, I used NEI 2017 and processed it with the EPA_ANTHRO_EMIS tool. Is there any modification needed in my "EPA_anthro_emis_2017NEI.inp"?
2, Did anyone get good PM2.5 CONUS simulation results with RADM2/MADE/SORGAM?
3, I appreciate any other suggestions.
Best,
Phillip