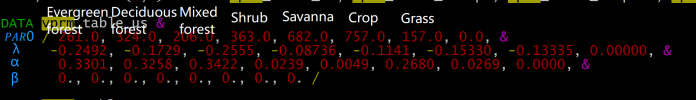
Hello everyone, I use the vprm parameter table when using the wrf vprm model, which contains many parameters, but I cannot know the specific format or order, which has caused me great trouble. If you are aware and willing to help me, I would greatly appreciate it.I have provided my understanding and speculation on the meaning of the parameter table below. Please be sure to let me know if it is correct. Thank you very much.
You are using an out of date browser. It may not display this or other websites correctly.
You should upgrade or use an alternative browser.
You should upgrade or use an alternative browser.
Questions about the vprm parameter table in wrf vprm
- Thread starter peng
- Start date
Hello everyone
I am now encountering a new problem when I try to modify chem/chemicals_ Vprm in init.F_ Table_ When using parameters, I recompiled WRF to make it effective, but encountered trouble and was unable to generate the four running files. However, when I used unmodified parameters, I successfully compiled, which caused great confusion for me. Do I need to recompile or modify the parameters of other files?
If you could provide some help or advice, I would greatly appreciate it! Below is my compilation and operation ./compile em_ real
I am now encountering a new problem when I try to modify chem/chemicals_ Vprm in init.F_ Table_ When using parameters, I recompiled WRF to make it effective, but encountered trouble and was unable to generate the four running files. However, when I used unmodified parameters, I successfully compiled, which caused great confusion for me. Do I need to recompile or modify the parameters of other files?
If you could provide some help or advice, I would greatly appreciate it! Below is my compilation and operation ./compile em_ real
Attachments
I found that after the modification of the compilation failed, I changed the value to the original way, and compiled again, and this time the compilation was successful, which made me very confused. I looked at the run log twice and there was a discrepancy around 332 lines. The one that failed didn't have -f90=gfortran. I'm pretty sure I entered my environment configuration after each compilation of./clean -a