Hello all!
The content of "ob.ascii" input to 4dvar is not reflected. We would like to know how to make it work effectively as an input file.
We have converted "ob.ascii" to input the observation results of the vertical Doppler LIDAR we installed as well as "ob.radar" as input files for 4dvar.
The format on pages 42 and 43 of the following document was used for the conversion.
We ran 4dvar, but the results did not appear to reflect the contents of "ob.ascii.
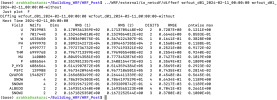
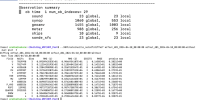
Checking the log "rsl.out.0000", it seems that 4dvar recognizes "ob.ascii" as an input file and reads "ob.ascii".
Therefore, I believe that the format of the converted "ob.ascii" is correct.
Attached are namelist.input, rsl.out.0000 and a reduced ob.ascii.
The "ob.ascii" has dummy values for testing.
Best regards.
The content of "ob.ascii" input to 4dvar is not reflected. We would like to know how to make it work effectively as an input file.
We have converted "ob.ascii" to input the observation results of the vertical Doppler LIDAR we installed as well as "ob.radar" as input files for 4dvar.
The format on pages 42 and 43 of the following document was used for the conversion.
We ran 4dvar, but the results did not appear to reflect the contents of "ob.ascii.
Checking the log "rsl.out.0000", it seems that 4dvar recognizes "ob.ascii" as an input file and reads "ob.ascii".
Therefore, I believe that the format of the converted "ob.ascii" is correct.
Attached are namelist.input, rsl.out.0000 and a reduced ob.ascii.
The "ob.ascii" has dummy values for testing.
Best regards.