Hi, friends
When I study ACI and set progn = 0, my WRF task stops.
but In my opinion if studing ACI, it must have aqueous phase chemistry.Because of the increased cloud water content, the aqueous phase chemistry is accelerated, which is also part of the ACI.
I just want to know how to solve it.
Here is my namelist.input
Here is my log of runing WRF-chem model.
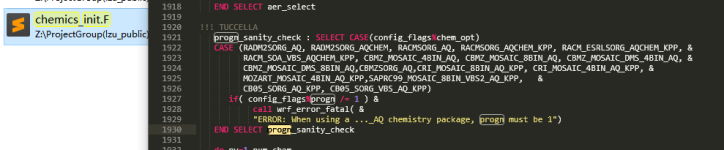

I got to set progn = 0, so that I can study ACI.
You can see that this mode was stopped midway.Is there something wrong with my namelist and code? I can't understand it ,and I hope someone could help me.Thank you for your message.
your friends,
cxyiyh
When I study ACI and set progn = 0, my WRF task stops.
but In my opinion if studing ACI, it must have aqueous phase chemistry.Because of the increased cloud water content, the aqueous phase chemistry is accelerated, which is also part of the ACI.
I just want to know how to solve it.
Here is my namelist.input
Code:
&physics
mp_physics = 10, 10, 10,
progn = 0, 0, 0,
ra_lw_physics = 4, 4, 4,
ra_sw_physics = 4, 4, 4,
radt = 30, 30, 30,
sf_sfclay_physics = 1, 1, 1,
sf_surface_physics = 3, 3, 3,
bl_pbl_physics = 1, 1, 1,
bldt = 0, 0, 0,
cu_physics = 3, 0, 0,
cu_diag = 1, 0, 0,
cudt = 5, 5, 5,
ishallow = 0,
isfflx = 1,
ifsnow = 1,
icloud = 1,
surface_input_source = 1,
num_soil_layers = 4,
sf_urban_physics = 0, 0, 0,
mp_zero_out = 2,
mp_zero_out_thresh = 1.e-12
maxiens = 1,
maxens = 3,
maxens2 = 3,
maxens3 = 16,
ensdim = 144,
cu_rad_feedback = .true.,
num_land_cat = 21,
/
&fdda
/
&dynamics
rk_ord = 3,
w_damping = 0,
diff_opt = 1, 1, 1,
km_opt = 4, 4, 4,
diff_6th_opt = 0, 0, 0,
diff_6th_factor = 0.12, 0.12, 0.12,
base_temp = 290.
damp_opt = 0,
zdamp = 5000., 5000., 5000.,
dampcoef = 0.2, 0.2, 0.2,
khdif = 0, 0, 0,
kvdif = 0, 0, 0,
non_hydrostatic = .true., .true., .true.,
moist_adv_opt = 2, 2, 2,
scalar_adv_opt = 2, 2, 2,
chem_adv_opt = 2, 2, 2,
tke_adv_opt = 2, 2, 2,
time_step_sound = 4, 4, 4,
h_mom_adv_order = 5, 5, 5,
v_mom_adv_order = 3, 3, 3,
h_sca_adv_order = 5, 5, 5,
v_sca_adv_order = 3, 3, 3,
/
&bdy_control
spec_bdy_width = 5,
spec_zone = 1,
relax_zone = 4,
specified = .true., .false., .false.,
nested = .false.,.true., .true.,
/
&grib2
/
&chem
kemit = 1,
chem_conv_tr = 1, 0, 0,
chem_opt = 34, 34, 34,
bioemdt = 30, 30, 30,
photdt = 30, 30, 30,
chemdt = 1.5, 1.5, 1.5,
io_style_emissions = 2,
emiss_opt = 5, 5, 5,
emiss_inpt_opt = 1, 1, 1,
emiss_opt_vol = 0, 0, 0,
emiss_ash_hgt = 20000.,
chem_in_opt = 0, 0, 0,
phot_opt = 1, 1, 1,
gas_drydep_opt = 1, 1, 1,
aer_drydep_opt = 301, 301, 301,
bio_emiss_opt = 0, 0, 0,
ne_area = 0,
dust_opt = 3,
dmsemis_opt = 1,
seas_opt = 1,
depo_fact = 0.25,
gas_bc_opt = 1, 1, 1,
gas_ic_opt = 1, 1, 1,
aer_bc_opt = 1, 1, 1,
aer_ic_opt = 1, 1, 1,
gaschem_onoff = 1, 1, 1,
aerchem_onoff = 1, 1, 1,
wetscav_onoff = 1, 1, 1,
cldchem_onoff = 0, 0, 0,
vertmix_onoff = 1, 1, 1,
conv_tr_wetscav = 1, 0, 0,
conv_tr_aqchem = 0, 0, 0,
biomass_burn_opt = 1, 1, 1,
plumerisefire_frq = 30, 30, 30,
have_bcs_chem = .false., .false., .false.,
aer_ra_feedback = 1, 1, 1,
aer_op_opt = 1, 1, 1,
opt_pars_out = 1,
diagnostic_chem = 0, 0, 0,
/
&namelist_quilt
nio_tasks_per_group = 0,
nio_groups = 1,
/Here is my log of runing WRF-chem model.
Code:
taskid: 0 hostname: n112
module_io_quilt_old.F 2931 F
Quilting with 1 groups of 0 I/O tasks.
Ntasks in X 5 , ntasks in Y 8
Domain # 1: dx = 27000.000 m
Domain # 2: dx = 9000.000 m
Domain # 3: dx = 3000.000 m
WRF V4.2 MODEL
*************************************
Parent domain
ids,ide,jds,jde 1 230 1 180
ims,ime,jms,jme -4 53 -4 30
ips,ipe,jps,jpe 1 46 1 23
*************************************
DYNAMICS OPTION: Eulerian Mass Coordinate
alloc_space_field: domain 1 , 370176348 bytes allocated
med_initialdata_input: calling input_input
Input data is acceptable to use: wrfinput_d01
CURRENT DATE = 2014-04-22_00:00:00
SIMULATION START DATE = 2014-04-22_00:00:00
Timing for processing wrfinput file (stream 0) for domain 1: 5.64336 elapsed seconds
Max map factor in domain 1 = 1.14. Scale the dt in the model accordingly.
D01: Time step = 90.00000 (s)
D01: Grid Distance = 27.00000 (km)
D01: Grid Distance Ratio dt/dx = 3.333333 (s/km)
D01: Ratio Including Maximum Map Factor = 3.792768 (s/km)
D01: NML defined reasonable_time_step_ratio = 6.000000
INPUT LandUse = "MODIFIED_IGBP_MODIS_NOAH"
LANDUSE TYPE = "MODIFIED_IGBP_MODIS_NOAH" FOUND 33 CATEGORIES 2 SEASONS WATER CATEGORY = 17 SNOW CATEGORY = 15
INITIALIZE THREE LSM RELATED TABLES
INPUT VEGPARM FOR MODI-RUC
VEGPARM FOR USGS FOUND 27 CATEGORIES
Skipping USGS table
VEGPARM FOR MODIFIED FOUND 20 CATEGORIES
Skipping MODIFIED table
VEGPARM FOR NLCD40 FOUND 40 CATEGORIES
Skipping NLCD40 table
VEGPARM FOR USGS-RUC FOUND 28 CATEGORIES
Skipping USGS-RUC table
VEGPARM FOR MODI-RUC FOUND 21 CATEGORIES
Found MODI-RUC table
Reading MODI-RUC table
INPUT SOIL TEXTURE CLASSIFICATION = STAS-RUC
SOIL TEXTURE CLASSIFICATION = STAS-RUC FOUND 19 CATEGORIES
*********************************************************************
* PROGRAM:WRF-Chem V4.2 MODEL
* *
* PLEASE REPORT ANY BUGS TO WRF-Chem HELP at *
* *
* wrfchemhelp.gsd@noaa.gov *
* *
*********************************************************************
WARNING: sea salt option 1 currently works only with the GOCART aerosol option.
call madronich phot initialization
output from subr init_data_mosaic_ptr
nphase_aer = 2
iphase = 1
ntype_aer = 1
ncomp_aer = 10
itype = 1
nsize_aer = 8
lptr_so4_aer 66 77 88 99 110 121 132 143
lptr_no3_aer 67 78 89 100 111 122 133 144
lptr_cl_aer 68 79 90 101 112 123 134 145
lptr_msa_aer 170 171 172 173 174 175 176 177
lptr_co3_aer 162 163 164 165 166 167 168 169
lptr_nh4_aer 69 80 91 102 113 124 135 146
lptr_na_aer 70 81 92 103 114 125 136 147
lptr_ca_aer 154 155 156 157 158 159 160 161
lptr_oin_aer 71 82 93 104 115 126 137 148
lptr_oc_aer 72 83 94 105 116 127 138 149
lptr_bc_aer 73 84 95 106 117 128 139 150
hyswptr_aer 74 85 96 107 118 129 140 151
waterptr_aer 75 86 97 108 119 130 141 152
numptr_aer 76 87 98 109 120 131 142 153
lptr_pcg1_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg2_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg3_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg4_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg5_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg6_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg7_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg8_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg9_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg1_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg2_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg3_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg4_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg5_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg6_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg7_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg8_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg9_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg1_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg2_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg3_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg4_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg5_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg6_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg7_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg8_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg1_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg2_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg3_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg4_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg5_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg6_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg7_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg8_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg1_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg2_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg3_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg4_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg5_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg6_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg7_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg8_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg9_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg1_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg2_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg3_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg4_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg5_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg6_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg7_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg8_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg9_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg1_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg2_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg3_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg4_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg5_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg6_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg7_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg8_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg1_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg2_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg3_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg4_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg5_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg6_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg7_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg8_f_o_aer 1 1 1 1 1 1 1 1
ant1_c_aer 1 1 1 1 1 1 1 1
ant2_c_aer 1 1 1 1 1 1 1 1
ant3_c_aer 1 1 1 1 1 1 1 1
ant4_c_aer 1 1 1 1 1 1 1 1
ant1_o_aer 1 1 1 1 1 1 1 1
ant2_o_aer 1 1 1 1 1 1 1 1
ant3_o_aer 1 1 1 1 1 1 1 1
ant4_o_aer 1 1 1 1 1 1 1 1
biog1_c_aer 1 1 1 1 1 1 1 1
biog2_c_aer 1 1 1 1 1 1 1 1
biog3_c_aer 1 1 1 1 1 1 1 1
biog4_c_aer 1 1 1 1 1 1 1 1
biog1_o_aer 1 1 1 1 1 1 1 1
biog2_o_aer 1 1 1 1 1 1 1 1
biog3_o_aer 1 1 1 1 1 1 1 1
biog4_o_aer 1 1 1 1 1 1 1 1
smpa_aer 1 1 1 1 1 1 1 1
smpbb_aer 1 1 1 1 1 1 1 1
glysoa_r1_aer 1 1 1 1 1 1 1 1
glysoa_r2_aer 1 1 1 1 1 1 1 1
glysoa_sfc_aer 1 1 1 1 1 1 1 1
glysoa_nh4_aer 1 1 1 1 1 1 1 1
glysoa_oh_aer 1 1 1 1 1 1 1 1
asoaX_aer 1 1 1 1 1 1 1 1
asoa1_aer 1 1 1 1 1 1 1 1
asoa2_aer 1 1 1 1 1 1 1 1
asoa3_aer 1 1 1 1 1 1 1 1
asoa4_aer 1 1 1 1 1 1 1 1
bsoaX_aer 1 1 1 1 1 1 1 1
bsoa1_aer 1 1 1 1 1 1 1 1
bsoa2_aer 1 1 1 1 1 1 1 1
bsoa3_aer 1 1 1 1 1 1 1 1
bsoa4_aer 1 1 1 1 1 1 1 1
sulfate 66 77 88 99 110 121 132 143 1
nitrate 67 78 89 100 111 122 133 144 2
chloride 68 79 90 101 112 123 134 145 3
carbonate 162 163 164 165 166 167 168 169 4
ammonium 69 80 91 102 113 124 135 146 5
sodium 70 81 92 103 114 125 136 147 6
calcium 154 155 156 157 158 159 160 161 7
otherinorg 71 82 93 104 115 126 137 148 8
organic-c 72 83 94 105 116 127 138 149 9
black-c 73 84 95 106 117 128 139 150 10
iphase = 2
ntype_aer = 1
ncomp_aer = 10
itype = 1
nsize_aer = 8
lptr_so4_aer 178 187 196 205 214 223 232 241
lptr_no3_aer 179 188 197 206 215 224 233 242
lptr_cl_aer 180 189 198 207 216 225 234 243
lptr_msa_aer 266 267 268 269 270 271 272 273
lptr_co3_aer 258 259 260 261 262 263 264 265
lptr_nh4_aer 181 190 199 208 217 226 235 244
lptr_na_aer 182 191 200 209 218 227 236 245
lptr_ca_aer 250 251 252 253 254 255 256 257
lptr_oin_aer 183 192 201 210 219 228 237 246
lptr_oc_aer 184 193 202 211 220 229 238 247
lptr_bc_aer 185 194 203 212 221 230 239 248
hyswptr_aer 74 85 96 107 118 129 140 151
waterptr_aer 75 86 97 108 119 130 141 152
numptr_aer 186 195 204 213 222 231 240 249
lptr_pcg1_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg2_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg3_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg4_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg5_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg6_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg7_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg8_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg9_b_c_aer 1 1 1 1 1 1 1 1
lptr_pcg1_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg2_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg3_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg4_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg5_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg6_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg7_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg8_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg9_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg1_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg2_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg3_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg4_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg5_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg6_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg7_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg8_b_c_aer 1 1 1 1 1 1 1 1
lptr_opcg1_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg2_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg3_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg4_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg5_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg6_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg7_b_o_aer 1 1 1 1 1 1 1 1
lptr_opcg8_b_o_aer 1 1 1 1 1 1 1 1
lptr_pcg1_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg2_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg3_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg4_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg5_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg6_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg7_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg8_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg9_f_c_aer 1 1 1 1 1 1 1 1
lptr_pcg1_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg2_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg3_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg4_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg5_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg6_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg7_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg8_f_o_aer 1 1 1 1 1 1 1 1
lptr_pcg9_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg1_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg2_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg3_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg4_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg5_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg6_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg7_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg8_f_c_aer 1 1 1 1 1 1 1 1
lptr_opcg1_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg2_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg3_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg4_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg5_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg6_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg7_f_o_aer 1 1 1 1 1 1 1 1
lptr_opcg8_f_o_aer 1 1 1 1 1 1 1 1
ant1_c_aer 1 1 1 1 1 1 1 1
ant2_c_aer 1 1 1 1 1 1 1 1
ant3_c_aer 1 1 1 1 1 1 1 1
ant4_c_aer 1 1 1 1 1 1 1 1
ant1_o_aer 1 1 1 1 1 1 1 1
ant2_o_aer 1 1 1 1 1 1 1 1
ant3_o_aer 1 1 1 1 1 1 1 1
ant4_o_aer 1 1 1 1 1 1 1 1
biog1_c_aer 1 1 1 1 1 1 1 1
biog2_c_aer 1 1 1 1 1 1 1 1
biog3_c_aer 1 1 1 1 1 1 1 1
biog4_c_aer 1 1 1 1 1 1 1 1
biog1_o_aer 1 1 1 1 1 1 1 1
biog2_o_aer 1 1 1 1 1 1 1 1
biog3_o_aer 1 1 1 1 1 1 1 1
biog4_o_aer 1 1 1 1 1 1 1 1
smpa_aer 1 1 1 1 1 1 1 1
smpbb_aer 1 1 1 1 1 1 1 1
glysoa_r1_aer 1 1 1 1 1 1 1 1
glysoa_r2_aer 1 1 1 1 1 1 1 1
glysoa_sfc_aer 1 1 1 1 1 1 1 1
glysoa_nh4_aer 1 1 1 1 1 1 1 1
glysoa_oh_aer 1 1 1 1 1 1 1 1
asoaX_aer 1 1 1 1 1 1 1 1
asoa1_aer 1 1 1 1 1 1 1 1
asoa2_aer 1 1 1 1 1 1 1 1
asoa3_aer 1 1 1 1 1 1 1 1
asoa4_aer 1 1 1 1 1 1 1 1
bsoaX_aer 1 1 1 1 1 1 1 1
bsoa1_aer 1 1 1 1 1 1 1 1
bsoa2_aer 1 1 1 1 1 1 1 1
bsoa3_aer 1 1 1 1 1 1 1 1
bsoa4_aer 1 1 1 1 1 1 1 1
sulfate 178 187 196 205 214 223 232 241 1
nitrate 179 188 197 206 215 224 233 242 2
chloride 180 189 198 207 216 225 234 243 3
carbonate 258 259 260 261 262 263 264 265 4
ammonium 181 190 199 208 217 226 235 244 5
sodium 182 191 200 209 218 227 236 245 6
calcium 250 251 252 253 254 255 256 257 7
otherinorg 183 192 201 210 219 228 237 246 8
organic-c 184 193 202 211 220 229 238 247 9
black-c 185 194 203 212 221 230 239 248 10
ltot, ltot2, lmaxd, l2maxd = 265 267 1200 1200
*** subr move_sections - method = 20
*** subr move_sections - idiag = 0
-------------- FATAL CALLED ---------------
FATAL CALLED FROM FILE: <stdin> LINE: 1958
ERROR: When using a ..._AQ chemistry package, progn must be 1
-------------------------------------------
Abort(1) on node 13 (rank 13 in comm 0): application called MPI_Abort(MPI_COMM_WORLD, 1) - process 13

I got to set progn = 0, so that I can study ACI.
You can see that this mode was stopped midway.Is there something wrong with my namelist and code? I can't understand it ,and I hope someone could help me.Thank you for your message.
your friends,
cxyiyh

