malimalihong
New member
hi everyone,
I'm using WRFchem4.3.3 to run a wrfchem case, but I found that as long as I use CBMZ related chemistry mechanism in the chemistry module, the mode doesn't run, it usually stops after 40-50 seconds of running, and it doesn't show in the rsl file error. But when I change the chemical mechanism to RADM2 (chem_opt=11) it works. Also, I run wrfchem3.9.1 with the same data and settings successfully. Does anyone know why?
This is the end of my rsl files and part of my nameless.input:
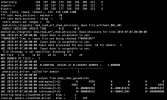
&time_control
run_days = 10,
run_hours = 0,
run_minutes = 0,
run_seconds = 0,
start_year = 2019, 2019, 2019,
start_month = 07, 07, 07,
start_day = 07, 07, 07,
start_hour = 00, 00, 00,
end_year = 2019, 2019, 2019,
end_month = 07, 07, 07,
end_day = 17, 17, 17,
end_hour = 00, 00, 00,
interval_seconds = 21600
input_from_file = .true.,.true.,.true.,
history_interval = 60, 60, 60,
frames_per_outfile = 1000, 1000, 1000,
restart = .false.,
restart_interval = 0,
io_form_history = 2,
io_form_restart = 2,
io_form_input = 2,
io_form_boundary = 2,
debug_level = 0,
io_form_auxinput5 = 2,
auxinput5_interval_m = 60, 60, 60,
/
&domains
time_step = 180,
time_step_fract_num = 0,
time_step_fract_den = 1,
max_dom = 3,
e_we = 100, 184, 136,
e_sn = 97, 178, 187,
e_vert = 33, 33, 33,
p_top_requested = 10000,
num_metgrid_levels = 38,
num_metgrid_soil_levels = 4,
dx = 36000, 12000, 4000,
dy = 36000, 12000, 4000,
grid_id = 1, 2, 3,
parent_id = 0, 1, 2,
i_parent_start = 1, 21, 69,
j_parent_start = 1, 18, 58,
parent_grid_ratio = 1, 3, 3,
parent_time_step_ratio = 1, 3, 3,
feedback = 1,
smooth_option = 0,
/
&physics
mp_physics = 2, 2, 2,
cu_physics = 5, 5, 0,
ra_lw_physics = 1, 1, 1,
ra_sw_physics = 1, 1, 1,
bl_pbl_physics = 2, 2, 2,
sf_sfclay_physics = 2, 2, 2,
sf_surface_physics = 2, 2, 2,
radt = 30, 30, 30,
bldt = 0, 0, 0,
cudt = 0, 0, 0,
isfflx = 1,
ifsnow = 0,
icloud = 1,
num_soil_layers = 4,
num_land_cat = 21,
sf_urban_physics = 0, 0, 0,
cu_diag = 1, 1, 0,
cu_rad_feedback = .true.,.false.,.false.,
progn = 1, 1, 1,
surface_input_source = 1,
sf_lake_physics = 1, 1, 1,
lakedepth_default = 8, 8, 8,
use_lakedepth = 1,
/
&fdda
grid_fdda = 1, 1, 1,
gfdda_inname = "wrffdda_d<domain>",
gfdda_end_h = 240, 240, 240,
gfdda_interval_m = 360, 360, 360,
fgdt = 0, 0, 0,
if_no_pbl_nudging_uv = 0, 0, 0,
if_no_pbl_nudging_t = 0, 0, 0,
if_no_pbl_nudging_q = 0, 0, 0,
if_zfac_uv = 0, 0, 0,
k_zfac_uv = 10, 10, 10,
if_zfac_t = 0, 0, 0,
k_zfac_t = 10, 10, 10,
if_zfac_q = 0, 0, 0,
k_zfac_q = 10, 10, 10,
guv = 0.0003, 0.0003, 0.0003,
gt = 0.0003, 0.0003, 0.0003,
gq = 0.0003, 0.0003, 0.0003,
if_ramping = 1,
dtramp_min = 60.0,
io_form_gfdda = 2,
/
&dynamics
w_damping = 0,
diff_opt = 1, 1, 1,
km_opt = 4, 4, 4,
diff_6th_opt = 0, 0, 0,
diff_6th_factor = 0.12, 0.12, 0.12,
base_temp = 290.
damp_opt = 0,
zdamp = 5000., 5000., 5000.,
dampcoef = 0.2, 0.2, 0.2,
khdif = 0, 0, 0,
kvdif = 0, 0, 0,
non_hydrostatic = .true., .true., .true.,
moist_adv_opt = 2, 2, 2,
scalar_adv_opt = 2, 2, 2,
chem_adv_opt = 2, 2, 2,
tke_adv_opt = 2, 2, 2,
gwd_opt = 0,
/
&bdy_control
spec_bdy_width = 5,
spec_zone = 1,
relax_zone = 4,
specified = .true., .false., .false.,
nested = .false., .true., .true.,
/
&grib2
/
&chem
kemit = 15,
chem_opt = 10, 10, 10,
bioemdt = 30, 30, 30,
photdt = 0, 0, 0,
chemdt = 0, 0, 0,
io_style_emissions = 1,
emiss_inpt_opt = 101, 101, 101,
emiss_opt = 3, 3, 15,
chem_in_opt = 0, 0, 0,
phot_opt = 2, 2, 2,
gas_drydep_opt = 1, 1, 1,
aer_drydep_opt = 1, 1, 1,
bio_emiss_opt = 0, 0, 0,
dust_opt = 0,
dmsemis_opt = 0,
seas_opt = 0,
gas_bc_opt = 1, 1, 1,
gas_ic_opt = 1, 1, 1,
aer_bc_opt = 1, 1, 1,
aer_ic_opt = 1, 1, 1,
gaschem_onoff = 1, 1, 1,
aerchem_onoff = 1, 1, 1,
wetscav_onoff = 1, 1, 1,
cldchem_onoff = 1, 1, 1,
vertmix_onoff = 1, 1, 1,
chem_conv_tr = 1, 1, 0,
biomass_burn_opt = 0, 0, 0,
plumerisefire_frq = 30, 30, 30,
aer_ra_feedback = 1, 1, 1,
have_bcs_chem = .false., .false., .false.,
aer_op_opt = 1, 1, 1,
opt_pars_out = 1,
/
&namelist_quilt
nio_tasks_per_group = 0,
nio_groups = 1,
/
I'm using WRFchem4.3.3 to run a wrfchem case, but I found that as long as I use CBMZ related chemistry mechanism in the chemistry module, the mode doesn't run, it usually stops after 40-50 seconds of running, and it doesn't show in the rsl file error. But when I change the chemical mechanism to RADM2 (chem_opt=11) it works. Also, I run wrfchem3.9.1 with the same data and settings successfully. Does anyone know why?
This is the end of my rsl files and part of my nameless.input:

&time_control
run_days = 10,
run_hours = 0,
run_minutes = 0,
run_seconds = 0,
start_year = 2019, 2019, 2019,
start_month = 07, 07, 07,
start_day = 07, 07, 07,
start_hour = 00, 00, 00,
end_year = 2019, 2019, 2019,
end_month = 07, 07, 07,
end_day = 17, 17, 17,
end_hour = 00, 00, 00,
interval_seconds = 21600
input_from_file = .true.,.true.,.true.,
history_interval = 60, 60, 60,
frames_per_outfile = 1000, 1000, 1000,
restart = .false.,
restart_interval = 0,
io_form_history = 2,
io_form_restart = 2,
io_form_input = 2,
io_form_boundary = 2,
debug_level = 0,
io_form_auxinput5 = 2,
auxinput5_interval_m = 60, 60, 60,
/
&domains
time_step = 180,
time_step_fract_num = 0,
time_step_fract_den = 1,
max_dom = 3,
e_we = 100, 184, 136,
e_sn = 97, 178, 187,
e_vert = 33, 33, 33,
p_top_requested = 10000,
num_metgrid_levels = 38,
num_metgrid_soil_levels = 4,
dx = 36000, 12000, 4000,
dy = 36000, 12000, 4000,
grid_id = 1, 2, 3,
parent_id = 0, 1, 2,
i_parent_start = 1, 21, 69,
j_parent_start = 1, 18, 58,
parent_grid_ratio = 1, 3, 3,
parent_time_step_ratio = 1, 3, 3,
feedback = 1,
smooth_option = 0,
/
&physics
mp_physics = 2, 2, 2,
cu_physics = 5, 5, 0,
ra_lw_physics = 1, 1, 1,
ra_sw_physics = 1, 1, 1,
bl_pbl_physics = 2, 2, 2,
sf_sfclay_physics = 2, 2, 2,
sf_surface_physics = 2, 2, 2,
radt = 30, 30, 30,
bldt = 0, 0, 0,
cudt = 0, 0, 0,
isfflx = 1,
ifsnow = 0,
icloud = 1,
num_soil_layers = 4,
num_land_cat = 21,
sf_urban_physics = 0, 0, 0,
cu_diag = 1, 1, 0,
cu_rad_feedback = .true.,.false.,.false.,
progn = 1, 1, 1,
surface_input_source = 1,
sf_lake_physics = 1, 1, 1,
lakedepth_default = 8, 8, 8,
use_lakedepth = 1,
/
&fdda
grid_fdda = 1, 1, 1,
gfdda_inname = "wrffdda_d<domain>",
gfdda_end_h = 240, 240, 240,
gfdda_interval_m = 360, 360, 360,
fgdt = 0, 0, 0,
if_no_pbl_nudging_uv = 0, 0, 0,
if_no_pbl_nudging_t = 0, 0, 0,
if_no_pbl_nudging_q = 0, 0, 0,
if_zfac_uv = 0, 0, 0,
k_zfac_uv = 10, 10, 10,
if_zfac_t = 0, 0, 0,
k_zfac_t = 10, 10, 10,
if_zfac_q = 0, 0, 0,
k_zfac_q = 10, 10, 10,
guv = 0.0003, 0.0003, 0.0003,
gt = 0.0003, 0.0003, 0.0003,
gq = 0.0003, 0.0003, 0.0003,
if_ramping = 1,
dtramp_min = 60.0,
io_form_gfdda = 2,
/
&dynamics
w_damping = 0,
diff_opt = 1, 1, 1,
km_opt = 4, 4, 4,
diff_6th_opt = 0, 0, 0,
diff_6th_factor = 0.12, 0.12, 0.12,
base_temp = 290.
damp_opt = 0,
zdamp = 5000., 5000., 5000.,
dampcoef = 0.2, 0.2, 0.2,
khdif = 0, 0, 0,
kvdif = 0, 0, 0,
non_hydrostatic = .true., .true., .true.,
moist_adv_opt = 2, 2, 2,
scalar_adv_opt = 2, 2, 2,
chem_adv_opt = 2, 2, 2,
tke_adv_opt = 2, 2, 2,
gwd_opt = 0,
/
&bdy_control
spec_bdy_width = 5,
spec_zone = 1,
relax_zone = 4,
specified = .true., .false., .false.,
nested = .false., .true., .true.,
/
&grib2
/
&chem
kemit = 15,
chem_opt = 10, 10, 10,
bioemdt = 30, 30, 30,
photdt = 0, 0, 0,
chemdt = 0, 0, 0,
io_style_emissions = 1,
emiss_inpt_opt = 101, 101, 101,
emiss_opt = 3, 3, 15,
chem_in_opt = 0, 0, 0,
phot_opt = 2, 2, 2,
gas_drydep_opt = 1, 1, 1,
aer_drydep_opt = 1, 1, 1,
bio_emiss_opt = 0, 0, 0,
dust_opt = 0,
dmsemis_opt = 0,
seas_opt = 0,
gas_bc_opt = 1, 1, 1,
gas_ic_opt = 1, 1, 1,
aer_bc_opt = 1, 1, 1,
aer_ic_opt = 1, 1, 1,
gaschem_onoff = 1, 1, 1,
aerchem_onoff = 1, 1, 1,
wetscav_onoff = 1, 1, 1,
cldchem_onoff = 1, 1, 1,
vertmix_onoff = 1, 1, 1,
chem_conv_tr = 1, 1, 0,
biomass_burn_opt = 0, 0, 0,
plumerisefire_frq = 30, 30, 30,
aer_ra_feedback = 1, 1, 1,
have_bcs_chem = .false., .false., .false.,
aer_op_opt = 1, 1, 1,
opt_pars_out = 1,
/
&namelist_quilt
nio_tasks_per_group = 0,
nio_groups = 1,
/
