You are using an out of date browser. It may not display this or other websites correctly.
You should upgrade or use an alternative browser.
You should upgrade or use an alternative browser.
convert_mpas error: forrtl: severe (174): SIGSEGV, segmentation fault occurred
- Thread starter Lucy
- Start date
The error message indicates that you don't have the file 'target_domain' available in your working directory.
Can you create this file and put it in the place where you run "conver_mpas" and try again?
Below is an example of 'target_domain':
nlat = 361
nlon = 720
startlat = -90.
startlon = -180.0
endlat = 90.
endlon = 180.0
By these options I remap global MPAS output to global 0.5 latitude x 0.5 longitude grids. You can change these options to remap your data to the grids you wants.
Can you create this file and put it in the place where you run "conver_mpas" and try again?
Below is an example of 'target_domain':
nlat = 361
nlon = 720
startlat = -90.
startlon = -180.0
endlat = 90.
endlon = 180.0
By these options I remap global MPAS output to global 0.5 latitude x 0.5 longitude grids. You can change these options to remap your data to the grids you wants.
It's perfectly fine to not have a 'target_domain' file -- as the printout says, "Default 0.5-degree global target domain will be used."
Just to verify that the "history" NetCDF file is valid, could you try running "ncdump -h history...nc" (where history...nc is the full name of your history file)?
If you haven't already done so, it might be worth trying to unlimit the stack size in your shell.
As a side note, in future, it's much better if you copy-and-paste error messages as text into your forum post. Images are not searchable on the forum, nor can anyone who is responding in a thread copy and paste text from the screenshot of the error messages.
Just to verify that the "history" NetCDF file is valid, could you try running "ncdump -h history...nc" (where history...nc is the full name of your history file)?
If you haven't already done so, it might be worth trying to unlimit the stack size in your shell.
As a side note, in future, it's much better if you copy-and-paste error messages as text into your forum post. Images are not searchable on the forum, nor can anyone who is responding in a thread copy and paste text from the screenshot of the error messages.
Thank you for your reply. I've tried "ncdump -h history.2014-09-15_00.00.00.nc"It's perfectly fine to not have a 'target_domain' file -- as the printout says, "Default 0.5-degree global target domain will be used."
Just to verify that the "history" NetCDF file is valid, could you try running "ncdump -h history...nc" (where history...nc is the full name of your history file)?
If you haven't already done so, it might be worth trying to unlimit the stack size in your shell.
As a side note, in future, it's much better if you copy-and-paste error messages as text into your forum post. Images are not searchable on the forum, nor can anyone who is responding in a thread copy and paste text from the screenshot of the error messages.
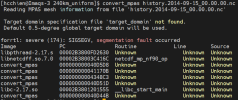
It seems it is valid (as the attatchment shown). I've also unlimited the stack size. However, the error message is still the same.
Reading MPAS mesh information from file 'history.2014-09-15_00.00.00.nc'
Target domain specification file 'target_domain' not found.
Default 0.5-degree global target domain will be used.
forrtl: severe (174): SIGSEGV, segmentation fault occurred
Image PC Routine Line Source
libpthread-2.17.s 00002B547F108630 Unknown Unknown Unknown
libnetcdff.so.7.0 00002B547E4FA16C netcdf_mp_nf90_op Unknown Unknown
convert_mpas 000000000040D5D8 Unknown Unknown Unknown
convert_mpas 000000000041170B Unknown Unknown Unknown
convert_mpas 00000000004348C3 Unknown Unknown Unknown
convert_mpas 000000000040D51D Unknown Unknown Unknown
libc-2.17.so 00002B547F337555 __libc_start_main Unknown Unknown
convert_mpas 000000000040D44B Unknown Unknown Unknown
P.S. Thank you for your suggestions, I will type the error message directly next time.
Attachments
razibvhuiyan911
New member
I struggle to find the exact information to set on target_domain, for a 60-3km mesh. For my case please guide me.The error message indicates that you don't have the file 'target_domain' available in your working directory.
Can you create this file and put it in the place where you run "conver_mpas" and try again?
Below is an example of 'target_domain':
nlat = 361
nlon = 720
startlat = -90.
startlon = -180.0
endlat = 90.
endlon = 180.0
By these options I remap global MPAS output to global 0.5 latitude x 0.5 longitude grids. You can change these options to remap your data to the grids you wants.
thank you
Razib
Razib,
Would you please take a look at the document here , which describes in detail how convert_mpas works and how to create 'target_domain'. You may also simply modify the sample 'target_domain' I posted above, then run convert_mpas.
Please let me know if you have any specific questions regarding this utility.
Would you please take a look at the document here , which describes in detail how convert_mpas works and how to create 'target_domain'. You may also simply modify the sample 'target_domain' I posted above, then run convert_mpas.
Please let me know if you have any specific questions regarding this utility.
I have a similar issue to Lucy above, this is my current setup:
San Diego Super Computer Center - Expanse
```
[cgrudzien@exp-18-46 test_data]$ nf-config --fc
/cm/shared/apps/spack/cpu/opt/spack/linux-centos8-zen2/intel-19.1.1.217/intel-mpi-2019.8.254-6kl5iseo7airrooh6wmqg2m3t5zvnsll/compilers_and_libraries_2020.2.254/linux/mpi/intel64/bin/mpiifort
[cgrudzien@exp-18-46 test_data]$ nf-config --fflags
-I/cm/shared/apps/spack/cpu/opt/spack/linux-centos8-zen2/intel-19.1.1.217/netcdf-fortran-4.5.3-u7d3te2y4gdabhq2yapkiwheqh2abavy/include -I/cm/shared/apps/spack/cpu/opt/spack/linux-centos8-zen2/intel-19.1.1.217/netcdf-fortran-4.5.3-u7d3te2y4gdabhq2yapkiwheqh2abavy/include
[cgrudzien@exp-18-46 test_data]$ nf-config --flibs
-L/cm/shared/apps/spack/cpu/opt/spack/linux-centos8-zen2/intel-19.1.1.217/netcdf-fortran-4.5.3-u7d3te2y4gdabhq2yapkiwheqh2abavy/lib -lnetcdff
```
I was able to successfully compile convert_mpas commit 248de60 with this setup
```
[cgrudzien@exp-18-46 test_data]$ convert_mpas
Usage: convert_mpas mesh-file [data-files]
If only one file argument is given, both the MPAS mesh information and
the fields will be read from the specified file.
If two or more file arguments are given, the MPAS mesh information will
be read from the first file and fields to be remapped will be read from
the subsequent files.
All time records from input files will be processed and appended to
the output file.
1
```
However, when I try to run this on a generic MPAS history file generated from a simulation with MPAS-A 8.0.1 on a shifted 60-3km variable resolution mesh, I get the following error message
```
Reading MPAS mesh information from file 'x20.835586.WestCoast.history.2022-12-23_00_00_00.nc'
Reading target domain specification from file 'target_domain'
Setting nlat = 361
Setting nlon = 720
Setting startlat = -90.00000
Setting startlon = -180.0000
Setting endlat = 90.00000
Setting endlon = 180.0000
forrtl: severe (174): SIGSEGV, segmentation fault occurred
Image PC Routine Line Source
convert_mpas 000000000044882A for__signal_handl Unknown Unknown
libpthread-2.28.s 00007FFFF5D48CF0 Unknown Unknown Unknown
libnetcdff.so.7.0 00007FFFF79BC117 netcdf_mp_nf90_ge Unknown Unknown
convert_mpas 000000000040CFD4 Unknown Unknown Unknown
convert_mpas 000000000041003A Unknown Unknown Unknown
convert_mpas 0000000000433095 Unknown Unknown Unknown
convert_mpas 000000000040C062 Unknown Unknown Unknown
libc-2.28.so 00007FFFF5629D85 __libc_start_main Unknown Unknown
convert_mpas 000000000040BF6E Unknown Unknown Unknown
```
A `ncdump -h` of the input file is included in the attached. Thanks for your help troubleshooting this.
San Diego Super Computer Center - Expanse
```
[cgrudzien@exp-18-46 test_data]$ nf-config --fc
/cm/shared/apps/spack/cpu/opt/spack/linux-centos8-zen2/intel-19.1.1.217/intel-mpi-2019.8.254-6kl5iseo7airrooh6wmqg2m3t5zvnsll/compilers_and_libraries_2020.2.254/linux/mpi/intel64/bin/mpiifort
[cgrudzien@exp-18-46 test_data]$ nf-config --fflags
-I/cm/shared/apps/spack/cpu/opt/spack/linux-centos8-zen2/intel-19.1.1.217/netcdf-fortran-4.5.3-u7d3te2y4gdabhq2yapkiwheqh2abavy/include -I/cm/shared/apps/spack/cpu/opt/spack/linux-centos8-zen2/intel-19.1.1.217/netcdf-fortran-4.5.3-u7d3te2y4gdabhq2yapkiwheqh2abavy/include
[cgrudzien@exp-18-46 test_data]$ nf-config --flibs
-L/cm/shared/apps/spack/cpu/opt/spack/linux-centos8-zen2/intel-19.1.1.217/netcdf-fortran-4.5.3-u7d3te2y4gdabhq2yapkiwheqh2abavy/lib -lnetcdff
```
I was able to successfully compile convert_mpas commit 248de60 with this setup
```
[cgrudzien@exp-18-46 test_data]$ convert_mpas
Usage: convert_mpas mesh-file [data-files]
If only one file argument is given, both the MPAS mesh information and
the fields will be read from the specified file.
If two or more file arguments are given, the MPAS mesh information will
be read from the first file and fields to be remapped will be read from
the subsequent files.
All time records from input files will be processed and appended to
the output file.
1
```
However, when I try to run this on a generic MPAS history file generated from a simulation with MPAS-A 8.0.1 on a shifted 60-3km variable resolution mesh, I get the following error message
```
Reading MPAS mesh information from file 'x20.835586.WestCoast.history.2022-12-23_00_00_00.nc'
Reading target domain specification from file 'target_domain'
Setting nlat = 361
Setting nlon = 720
Setting startlat = -90.00000
Setting startlon = -180.0000
Setting endlat = 90.00000
Setting endlon = 180.0000
forrtl: severe (174): SIGSEGV, segmentation fault occurred
Image PC Routine Line Source
convert_mpas 000000000044882A for__signal_handl Unknown Unknown
libpthread-2.28.s 00007FFFF5D48CF0 Unknown Unknown Unknown
libnetcdff.so.7.0 00007FFFF79BC117 netcdf_mp_nf90_ge Unknown Unknown
convert_mpas 000000000040CFD4 Unknown Unknown Unknown
convert_mpas 000000000041003A Unknown Unknown Unknown
convert_mpas 0000000000433095 Unknown Unknown Unknown
convert_mpas 000000000040C062 Unknown Unknown Unknown
libc-2.28.so 00007FFFF5629D85 __libc_start_main Unknown Unknown
convert_mpas 000000000040BF6E Unknown Unknown Unknown
```
A `ncdump -h` of the input file is included in the attached. Thanks for your help troubleshooting this.
Attachments
Investigating further, I think this was just a memory issue, I was able to get this to work by using additional memory / reducing the number of fields that I was interpolatingI have a similar issue to Lucy above, this is my current setup:
San Diego Super Computer Center - Expanse
```
[cgrudzien@exp-18-46 test_data]$ nf-config --fc
/cm/shared/apps/spack/cpu/opt/spack/linux-centos8-zen2/intel-19.1.1.217/intel-mpi-2019.8.254-6kl5iseo7airrooh6wmqg2m3t5zvnsll/compilers_and_libraries_2020.2.254/linux/mpi/intel64/bin/mpiifort
[cgrudzien@exp-18-46 test_data]$ nf-config --fflags
-I/cm/shared/apps/spack/cpu/opt/spack/linux-centos8-zen2/intel-19.1.1.217/netcdf-fortran-4.5.3-u7d3te2y4gdabhq2yapkiwheqh2abavy/include -I/cm/shared/apps/spack/cpu/opt/spack/linux-centos8-zen2/intel-19.1.1.217/netcdf-fortran-4.5.3-u7d3te2y4gdabhq2yapkiwheqh2abavy/include
[cgrudzien@exp-18-46 test_data]$ nf-config --flibs
-L/cm/shared/apps/spack/cpu/opt/spack/linux-centos8-zen2/intel-19.1.1.217/netcdf-fortran-4.5.3-u7d3te2y4gdabhq2yapkiwheqh2abavy/lib -lnetcdff
```
I was able to successfully compile convert_mpas commit 248de60 with this setup
```
[cgrudzien@exp-18-46 test_data]$ convert_mpas
Usage: convert_mpas mesh-file [data-files]
If only one file argument is given, both the MPAS mesh information and
the fields will be read from the specified file.
If two or more file arguments are given, the MPAS mesh information will
be read from the first file and fields to be remapped will be read from
the subsequent files.
All time records from input files will be processed and appended to
the output file.
1
```
However, when I try to run this on a generic MPAS history file generated from a simulation with MPAS-A 8.0.1 on a shifted 60-3km variable resolution mesh, I get the following error message
```
Reading MPAS mesh information from file 'x20.835586.WestCoast.history.2022-12-23_00_00_00.nc'
Reading target domain specification from file 'target_domain'
Setting nlat = 361
Setting nlon = 720
Setting startlat = -90.00000
Setting startlon = -180.0000
Setting endlat = 90.00000
Setting endlon = 180.0000
forrtl: severe (174): SIGSEGV, segmentation fault occurred
Image PC Routine Line Source
convert_mpas 000000000044882A for__signal_handl Unknown Unknown
libpthread-2.28.s 00007FFFF5D48CF0 Unknown Unknown Unknown
libnetcdff.so.7.0 00007FFFF79BC117 netcdf_mp_nf90_ge Unknown Unknown
convert_mpas 000000000040CFD4 Unknown Unknown Unknown
convert_mpas 000000000041003A Unknown Unknown Unknown
convert_mpas 0000000000433095 Unknown Unknown Unknown
convert_mpas 000000000040C062 Unknown Unknown Unknown
libc-2.28.so 00007FFFF5629D85 __libc_start_main Unknown Unknown
convert_mpas 000000000040BF6E Unknown Unknown Unknown
```
A `ncdump -h` of the input file is included in the attached. Thanks for your help troubleshooting this.
```
Reading MPAS mesh information from file 'x20.835586.WestCoast.history.2022-12-2
3_00_00_00.nc'
Reading target domain specification from file 'target_domain'
Setting nlat = 361
Setting nlon = 720
Setting startlat = -90.00000
Setting startlon = -180.0000
Setting endlat = 90.00000
Setting endlon = 180.0000
Computing remapping weights
Time to compute remap weights: 1.659505 s
Created a new output file
Reading list of fields to include in remapped output
Found 1 lines in 'include_fields' file
List of fields to be excluded from output ('exclude_fields') not found
Remapping MPAS fields from file 'x20.835586.WestCoast.history.2022-12-23_00_00_
00.nc'
Input file has 1 records
Defining fields in output file
Remapping field rainc, frame 1
read: 0.005019 s
remap: 0.002869 s
write: 0.000445 s
Total runtime: 5.476796
```
Last edited:
Thanks for the response, indeed, it seems like was not memory related. I'm able to run all fields through convert_mpas in a batch job but not an interactive job on a compute node, most likely due to local settings on our HPC system. Interestingly, this issue does not come up when running convert_mpas in a singularity container, only when I am running the version I built versus the system modules. That is to say, I can run convert_mpas in either a batch or interactive job from the container, but only from a batch job when using the system version I built. We're still debugging to determine the source of the issue.
Getting deeper into this, it looks like there is an issue specifically with how our `salloc` sessions and our system libraries are interacting on Expanse. I tested building convert_mpas and all of its dependences from scratch on the system using only a gcc module load and loosely following my singularity container definition. The build was successful, and the convert_mpas executable I built with dependencies built from scratch runs in an interactive session while the build from the system library seg faults.