AndrewLowry
New member
I am hoping someone can help me with a model crash that occurred after 24 years of simulation. A bit of background, it is a paleo climate simulation for 12k BP using WRF v.4.4. I have modified the phys files rrtmg_lw, rrtmg_sw, radiation_driver to account for paleo orbital parameters and GHG values. I also modified module_cu_bmj to revert FSS and FSL=0.6 as per Fonseca et al. 2015. For WPS I have provided my own geogrid binary files for LANDUSEF, HGT_M, ALBEDO12M, LAI12M, GREENFRAC12M, SOILTEMP. I've attached the namelist.input showing all the phys etc. used and namelist.wps.
I have diag_print turned on, and attach a truncated part of the rsl.error.0000 file that includes initial details and then the last period where the model crashed (it'd be too big otherwise). It shows dmudt and dpsdt becoming nan followed by sfcevp, hfx, lh in the following timestep and thereafter segmentation fault issues. The model started failing at date 0065-04-19_18:54, so I looked at the metgrid file for 18:00 of that day and did not find anything suspicious nor was there anything in wrflowinp_d01.
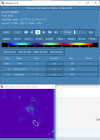
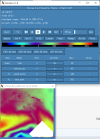
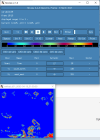
I looked through the wrfout file at the last timestep (0065-04-19_19:00). The thing that looks odd is that GRDFLX has 4 grid cells that are nan (screenshot attached) and there is a collection of 3D variables that have an empty patch: ACHFX, ACLHF, DUSFCG, DVSFCG, HFX, LH, MU, PBLH, PSFC, T2, TH2, U10, U10E, UST, V10, V10E; and the following 4D variables: DTAUX3D, DTAUY3D, EL_PBL (only level 3 and upwards), P, PH (only level 2 and upwards), P_HYD, QNCCN, QVAPOR, T, THM, TKE_PBL, U, V, W. The patch for MU (3D) and P (4D) is larger than the others. I've attached a few sample ncview screenshots of what this looks like. There was also a very interesting rectangle, but not nan values, in the variables NOAHRES and in the same spot in SR, also attached.
I don't really know what is going on, it seems unlikely to be related to my phys modifications as they succeeded for 24 years. My suspicion is it is either something in the input file (grib files I created from CESM output). Or more likely it is the GRDFLX which propogated to the other variables. The location of these cells is the southern tip of New Zealand and is likely they were very snowy, possibly glacial, but not categorised as such in the appropriate variables. These are just guesses and I would love some help to diagnose the problem. Please let me know what else I can attach that will help as well.
I have diag_print turned on, and attach a truncated part of the rsl.error.0000 file that includes initial details and then the last period where the model crashed (it'd be too big otherwise). It shows dmudt and dpsdt becoming nan followed by sfcevp, hfx, lh in the following timestep and thereafter segmentation fault issues. The model started failing at date 0065-04-19_18:54, so I looked at the metgrid file for 18:00 of that day and did not find anything suspicious nor was there anything in wrflowinp_d01.
I looked through the wrfout file at the last timestep (0065-04-19_19:00). The thing that looks odd is that GRDFLX has 4 grid cells that are nan (screenshot attached) and there is a collection of 3D variables that have an empty patch: ACHFX, ACLHF, DUSFCG, DVSFCG, HFX, LH, MU, PBLH, PSFC, T2, TH2, U10, U10E, UST, V10, V10E; and the following 4D variables: DTAUX3D, DTAUY3D, EL_PBL (only level 3 and upwards), P, PH (only level 2 and upwards), P_HYD, QNCCN, QVAPOR, T, THM, TKE_PBL, U, V, W. The patch for MU (3D) and P (4D) is larger than the others. I've attached a few sample ncview screenshots of what this looks like. There was also a very interesting rectangle, but not nan values, in the variables NOAHRES and in the same spot in SR, also attached.
I don't really know what is going on, it seems unlikely to be related to my phys modifications as they succeeded for 24 years. My suspicion is it is either something in the input file (grib files I created from CESM output). Or more likely it is the GRDFLX which propogated to the other variables. The location of these cells is the southern tip of New Zealand and is likely they were very snowy, possibly glacial, but not categorised as such in the appropriate variables. These are just guesses and I would love some help to diagnose the problem. Please let me know what else I can attach that will help as well.
Attachments
-
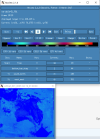 EL_PBL_lvl2.PNG60.8 KB · Views: 6
EL_PBL_lvl2.PNG60.8 KB · Views: 6 -
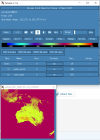 GRDFLX.PNG47.9 KB · Views: 4
GRDFLX.PNG47.9 KB · Views: 4 -
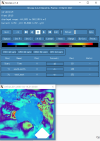 LH.png88.6 KB · Views: 3
LH.png88.6 KB · Views: 3 -
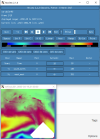 MU.png60.6 KB · Views: 3
MU.png60.6 KB · Views: 3 -
namelist.input.txt6 KB · Views: 8
-
namelist.wps.txt837 bytes · Views: 2
-
 NOAHRES.PNG35.7 KB · Views: 3
NOAHRES.PNG35.7 KB · Views: 3 -
 P.PNG57 KB · Views: 3
P.PNG57 KB · Views: 3 -
rsl.error.0000.txt179.3 KB · Views: 1
-
 SR.PNG48.2 KB · Views: 4
SR.PNG48.2 KB · Views: 4
