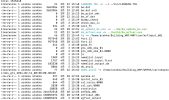
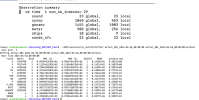
./da_wrfvar.exe error:
module_io_quilt_old.F 2931 T
Namelist logging not found in namelist.input. Using registry defaults for variables in logging.
*** VARIATIONAL ANALYSIS ***
WRFDA V4.5.2
Ntasks in X 10, ntasks in Y 10
*************************************
Parent domain
ids,ide,jds,jde 1 304 1 294
ims,ime,jms,jme -4 38 -4 37
ips,ipe,jps,jpe 1 31 1 30
*************************************
DYNAMICS OPTION: Eulerian Mass Coordinate
alloc_space_field: domain 1, 86418536 bytes allocated
Input data is acceptable to use: fg
hybrid_opt = 2
use_theta_m = 1
WRF NUMBER OF TILES FROM OMP_GET_MAX_THREADS = 36
Tile Strategy is not specified. Assuming 1D-Y
....
...
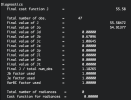
Domain mapping info:
map_proj = 3
cen_lat = 0.225000E+02
cen_lon = 0.865000E+02
truelat1 = 0.225000E+02
truelat2 = 0.000000E+00
start_lat = 0.171538E+02
start_lon = 0.806196E+02
pole_lat = 0.900000E+02
dsm = 0.400000E+01
module_io_quilt_old.F 2931 T
Namelist logging not found in namelist.input. Using registry defaults for variables in logging.
*** VARIATIONAL ANALYSIS ***
WRFDA V4.5.2
Ntasks in X 10, ntasks in Y 10
*************************************
Parent domain
ids,ide,jds,jde 1 304 1 294
ims,ime,jms,jme -4 38 -4 37
ips,ipe,jps,jpe 1 31 1 30
*************************************
DYNAMICS OPTION: Eulerian Mass Coordinate
alloc_space_field: domain 1, 86418536 bytes allocated
Input data is acceptable to use: fg
hybrid_opt = 2
use_theta_m = 1
WRF NUMBER OF TILES FROM OMP_GET_MAX_THREADS = 36
Tile Strategy is not specified. Assuming 1D-Y
....
...
Domain mapping info:
map_proj = 3
cen_lat = 0.225000E+02
cen_lon = 0.865000E+02
truelat1 = 0.225000E+02
truelat2 = 0.000000E+00
start_lat = 0.171538E+02
start_lon = 0.806196E+02
pole_lat = 0.900000E+02
dsm = 0.400000E+01
Last edited: