V4.4
No git found or not a git repository, git commit version not available.
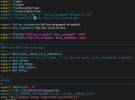
Compiling: WRF_EM_CORE
Linux Archangel11 5.15.153.1-microsoft-standard-WSL2 #1 SMP Fri Mar 29 23:14:13 UTC 2024 x86_64 x86_64 x86_64 GNU/Linux
GNU Fortran (Ubuntu 11.4.0-1ubuntu1~22.04) 11.4.0
Copyright (C) 2021 Free Software Foundation, Inc.
This is free software; see the source for copying conditions. There is NO
warranty; not even for MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE.
==============================================================================================
setting parallel make -j 2
==============================================================================================
The following indicate the compilers selected to build the WRF system
Serial Fortran compiler (mostly for tool generation):
which SFC
/usr/bin/gfortran
Serial C compiler (mostly for tool generation):
which SCC
/usr/bin/gcc
Fortran compiler for the model source code:
which FC
/home/ioga1101/install/mpich/bin/mpif90
Will use 'time' to report timing information
C compiler for the model source code:
which CC
/home/ioga1101/install/mpich/bin/mpicc
==============================================================================================
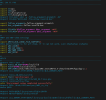
make -i -r MODULE_DIRS="-I../dyn_em -I/home/ioga1101/chem/WRFV4.4/external/esmf_time_f90 -I/home/ioga1101/chem/WRFV4.4/main -I/home/ioga1101/chem/WRFV4.4/external/io_netcdf -I/home/ioga1101/chem/WRFV4.4/external/io_int -I/home/ioga1101/chem/WRFV4.4/frame -I/home/ioga1101/chem/WRFV4.4/share -I/home/ioga1101/chem/WRFV4.4/phys -I/home/ioga1101/chem/WRFV4.4/wrftladj -I/home/ioga1101/chem/WRFV4.4/chem -I/home/ioga1101/chem/WRFV4.4/inc -I/home/ioga1101/install/netcdf/include " ext
make[1]: Entering directory '/home/ioga1101/chem/WRFV4.4'
--------------------------------------
if [ 0 -eq 0 ] ; then \
( cd frame ; make -i -r externals ) ; \
else \
( cd frame ; make -i -r PLUSFLAG="-DWRFPLUS=1" externals ) ; \
fi
make[2]: Entering directory '/home/ioga1101/chem/WRFV4.4/frame'
( cd /home/ioga1101/chem/WRFV4.4/external/esmf_time_f90 ; \
make -j 2 FC="gfortran -w -ffree-form -ffree-line-length-none -fconvert=big-endian -frecord-marker=4 -fallow-argument-mismatch -fallow-invalid-boz " RANLIB="ranlib" \
CPP="/lib/cpp -P -nostdinc -I/home/ioga1101/chem/WRFV4.4/inc -I. -DEM_CORE=1 -DNMM_CORE=0 -DNMM_MAX_DIM=2600 -DDA_CORE=0 -DWRFPLUS=0 -DIWORDSIZE=4 -DDWORDSIZE=8 -DRWORDSIZE=4 -DLWORDSIZE=4 -DNONSTANDARD_SYSTEM_SUBR -DWRF_USE_CLM -DUSE_NETCDF4_FEATURES -DWRFIO_NCD_LARGE_FILE_SUPPORT -DDM_PARALLEL -DNETCDF -DLANDREAD_STUB=1 -DUSE_ALLOCATABLES -Dwrfmodel -DGRIB1 -DINTIO -DKEEP_INT_AROUND -DLIMIT_ARGS -DBUILD_RRTMG_FAST=0 -DBUILD_RRTMK=0 -DBUILD_SBM_FAST=1 -DSHOW_ALL_VARS_USED=0 -DCONFIG_BUF_LEN=65536 -DMAX_DOMAINS_F=21 -DMAX_HISTORY=25 -DNMM_NEST=0 -traditional-cpp -DUSE_NETCDF4_FEATURES -DWRFIO_NCD_LARGE_FILE_SUPPORT" AR="ar" ARFLAGS="ru" )
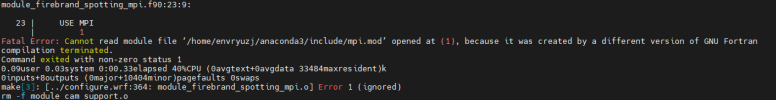
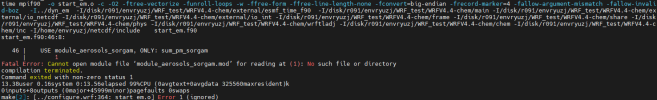
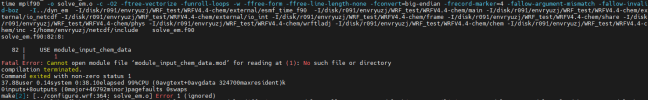
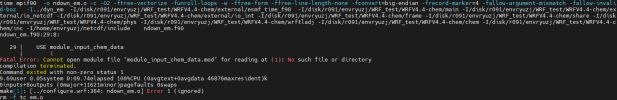
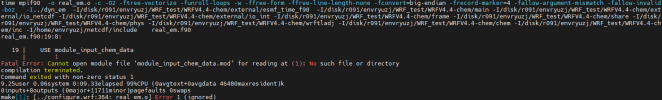
at this part we got some different build, so before continue with chem installation, can i see all your libraries version also how you configure your libraries,
Hi, Thanks for reply, I list all the libraries here, hdf5-1.10.5, flex-2.6.4, jasper-1.900.1,libpng-1.2.50, libtirpc-1.3.3, mpich-3.0.4, netcdf-4.1.3, zlib-1.2.7
The env setting is in below:
export WRF_DIR=/disk/r091/envryuzj/WRF_test/WRFV44
#NETCDF setting
#------------------------------------------------------------------------------------------
export NETCDF=$HOME/netcdf
export PATH=$HOME/netcdf/bin:$PATH
#export PATH=/home/envryuzj/netcdf/bin:$PATH
export NETCDF_LIB=$NETCDF/lib
export NETCDF_INC=$NETCDF/include
export NETCDF_DIR=$NETCDF
export LD_LIBRARY_PATH=$NETCDF/lib:$LD_LIBRARY_PATH
export NETCDF_classic=1
#cd /disk/r091/envryuzj/WRF_test/WRFV4.4
#/home/envryuzj/netcdf/bin/nc-config --all
#------------------------------------------------------------------------------------------
#mpich
#------------------------------------------------------------------------------------------
#export MPICH=$HOME/mpich/bin:$PATH
export PATH=$MPICH/bin:$PATH
#export MPICH_LIB=$MPICH/lib
#export MPICH_INC=$MPICH/include
export LD_LIBRARY_PATH=$MPICH/lib:$LD_LIBRARY_PATH
#------------------------------------------------------------------------------------------
#zlib
#------------------------------------------------------------------------------------------
export ZLIB_HOME=$HOME/zlib
export LD_LIBRARY_PATH=$ZLIB_HOME/lib:$LD_LIBRARY_PATH
#export ZLIB=$HOME/zlib/bin:$PATH
#export PATH=$ZLIB_HOME/bin:$PATH
#export ZLIB_LIB=$ZLIB/lib
#export ZLIB_INC=$ZLIB/include
#export LD_LIBRARY_PATH=$ZLIB/lib:$LD_LIBRARY_PATH
#------------------------------------------------------------------------------------------
#hdf5
#------------------------------------------------------------------------------------------
#export CPPFLAGS=-I$HOME/hdf5/include
#export LDFLAGS=-L$HOME/hdf5/lib
#export LD_LIBRARY_PATH=$HOME/hdf5/lib
export HDF5=$HOME/hdf5
export PATH=/home/envryuzj/hdf5/bin:$PATH
#export HDF5_LIB=$HDF5/lib
#export HDF5_INC=$HDF5/include
export LD_LIBRARY_PATH=$HDF5/lib:$LD_LIBRARY_PATH
# libpng---------------------------------------------------------------------------------
export PNG_LIB=$HOME/libpng
#export PATH=$PNG_LIB/bin:$PATH
export PNG_INC=$PNG_LIB/include
#export LD_LIBRARY_PATH=$PNG_LIB/lib:$LD_LIBRARY_PATH
# gssapi---------------------------------------------------------------------------------
export GSSAPI_LIB=$HOME/libtirpc
#export PATH=$GSSAPI_LIB/bin:$PATH
export GSSAPI_INC=$GSSAPI_LIB/include
#export LD_LIBRARY_PATH=$GSSAPI_LIB/lib:$LD_LIBRARY_PATH
#------------------------------------------------------------------------------------------
#JASPER
#------------------------------------------------------------------------------------------
export JASPER=$HOME/jasper
#export PATH=$JASPER/bin:$PATH
export JASPERLIB=$JASPER/lib
export JASPERINC=$JASPER/include/jasper
export LD_LIBRARY_PATH=$JASPERLIB:$LD_LIBRARY_PATH
export CPPFLAGS="-I$JASPERINC"
export LDFLAGS="-L$JASPERLIB -ljasper"
echo "Jasper setup completed successfully"
#------------------------------------------------------------------------------------------
#CC CXX FC F90
#------------------------------------------------------------------------------------------
export CC=gcc
export CXX=g++
export FC=/bin/gfortran
export F77=/bin/gfortran
#export FCFLAGS="-m64 -w -fallow-argument-mismatch -O2"
#export FFLAGS="$FCFLAGS -fno-allow-invalid-boz"
export fallow_argument=-fallow-argument-mismatch
export boz_argument=-fallow-invalid-boz
export FFLAGS="$fallow_argument $boz_argument -m64"
export FCFLAGS="$fallow_argument $boz_argument -m64"
#-----------------------------------------------------------------------------------------
#WRF and WRF-Chem settings
#------------------------------------------------------------------------------------------
export WRFIO_NCD_LARGE_FILE_SUPPORT=1
#the modify below is for wrf-chem, if it can not work, just zhushudiao jiuhaole
export WRF_CHEM=1
export WRF_EM_CORE=1
export WRF_NMM_CORE=0
export WRF_KPP=1
#------------------------------------------------------------------------------------------
#FLEX
#------------------------------------------------------------------------------------------
#export YACC='yacc -d'
export FLEX_LIB_DIR=$HOME/local/lib
export FLEX=$HOME/local/bin/flex
export KPP_HOME=/disk/r091/envryuzj/WRF_test/WRFV4.4-chem/chem/KPP/kpp/kpp-2.1
#export LD_LIBRARY_PATH=$HOME/local/lib:$LD_LIBRARY_PATH
#export FLEX_LIB_DIR=/usr/lib
export YACC='/usr/bin/yacc -d'
#export FLEX_LIB_DIR=/usr/lib/x86_64-linux-gnu
#export FLEX=/usr/bin/flex
echo "Environment setup completed successfully"