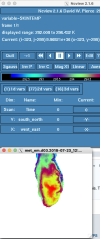
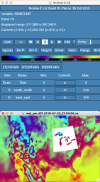
Hi all,
I'm in the process of updating SSTs in the WRF v3.8 model but I'm running into WPS issues when attempting to convert GLSEA SST data from netcdf to grib. I'm aware that there are similar posts on this forum however, after trying out a couple of different options, I am still running into issues with this conversion.
So far I have attempted to use the iris package in python to convert netcdf to grib which results in Grib code '255' ie missing

I've also tried to convert the files using cdo following this tutorial. While I manage to produce grib files, ungrib results in empty files. I have attempted to use sst parameter ids (id 34) and skin temp (id 235) codes from ECMWF tables which produce grib files (logs below) however, ungrib is unsuccessful.

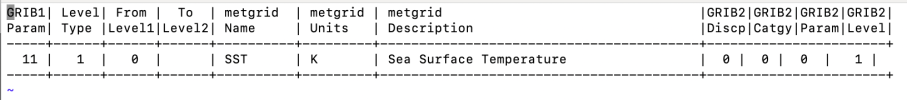
I am very unfamiliar with grib file manipulation and using my limited understanding, I assumed that if my Grib file had similar codes (e.g. 235) which matched the Vtable is WPS then ungrib should work, which is not the case (Vtable.ECMWF and generic Vtable.SST screenshots below).

Any help to adequately convert netcdf to grib would be greatly appreciated.
I'm in the process of updating SSTs in the WRF v3.8 model but I'm running into WPS issues when attempting to convert GLSEA SST data from netcdf to grib. I'm aware that there are similar posts on this forum however, after trying out a couple of different options, I am still running into issues with this conversion.
So far I have attempted to use the iris package in python to convert netcdf to grib which results in Grib code '255' ie missing

I've also tried to convert the files using cdo following this tutorial. While I manage to produce grib files, ungrib results in empty files. I have attempted to use sst parameter ids (id 34) and skin temp (id 235) codes from ECMWF tables which produce grib files (logs below) however, ungrib is unsuccessful.

I am very unfamiliar with grib file manipulation and using my limited understanding, I assumed that if my Grib file had similar codes (e.g. 235) which matched the Vtable is WPS then ungrib should work, which is not the case (Vtable.ECMWF and generic Vtable.SST screenshots below).


Any help to adequately convert netcdf to grib would be greatly appreciated.